Introduction
Authors: Andy Bunn  , Ed Cook, Kevin Anchukaitis
, Ed Cook, Kevin Anchukaitis  , Tyson Lee Swetnam
, Tyson Lee Swetnam 
Developers: Ifeoluwa Ale  , Michele Cosi
, Michele Cosi 

Welcome¶
Welcome to openDendro, the home of an open-source framework for the essential analytic software used in dendrochronology. Our aims are to provide researchers working with tree-ring data the necessary tools in open-source environments, and to promote open science practices and enhance the rigor and transparency in dendrochronological research.
At openDendro, we recognize the importance of the foundational scientific software in advancing knowledge, enabling collaboration, ensuring reproducibility, and contributing to the collective progress of the dendrochronology community.
Our framework seamlessly integrates with popular programming languages R and Python with public research data. We work to accommodate diverse researchers' preferences and workflows while embracing FAIR, CARE, and TRUST data principles in the effort to foster innovation and facilitate the sharing of knowledge within the field of dendrochronology and its many trans-disciplinary applications.
History and Motivation¶
For decades, much of the software used in dendrochronology has been in legacy programming languages which have been maintained by a small number of volunteers. These codebases are at risk of becoming inoperable or incompatible as advances in computing architecture accelerate. This is colloquially known as the "Some random person in Nebraska" problem in homage to a cartoon from xckd. And that random person is Ed Cook of course.
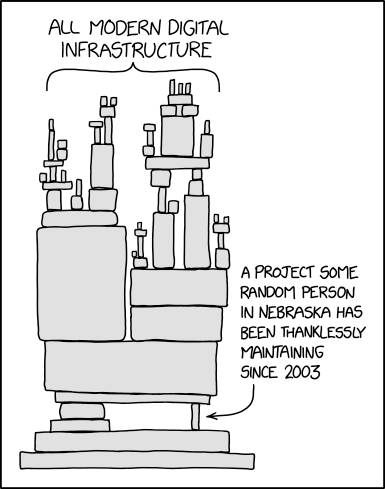
Our motivation with this project is to develop and foster a community of developers to maintain and build open-source software that will be usable by researchers who use tree-ring data. Modernizing and enhancing this software will also extend both their reach and utility beyond the dendrochronology community and allow integration into related initiatives in the atmospheric and earth sciences. We hope that making these tools open source will also facilitate further collaborative development, broaden the responsibility for collective maintenance and enhancement of this software, and ensure the persistence of these tools.
Why Write Code?¶
Writing code in languages like Python or R makes reproducible science easier compared to GUI-based analysis. Scripting offers transparency, automation, replication, portability, extensibility, and version control, which enhance reproducibility in scientific research.
-
Transparency and Documentation: Scripts are plain-text files that document the analysis workflow, making it easier for other researchers to understand and reproduce the analysis. GUI-based analysis lacks clear documentation of the steps and parameters used.
-
Automation and Efficiency: Scripts automate repetitive tasks, saving time and reducing human errors. Code snippets, functions, and libraries can be reused across multiple datasets or scenarios, ensuring efficient and consistent analysis.
-
Replication: Scripts facilitate straightforward replication of analyses. Other researchers can run the script on their own machines with the same input data and obtain identical results. GUI-based analysis often requires recreating specific interactions and settings, which is error-prone and time-consuming.
-
Portability and Compatibility: Scripts in Python or R are portable and can run on different platforms without significant modifications. GUI-based tools may have platform-specific dependencies or version requirements, limiting their portability and compatibility.
-
Extensibility and Flexibility: Scripting languages provide a wide range of libraries, packages, and modules, enabling advanced analyses, integration with external tools, and customization. GUI-based tools often have limited built-in functionality, making extension and adaptation challenging.
-
Version Control: Scripts can easily integrate with version control systems like Git, allowing researchers to track changes, collaborate, and maintain an analysis history. Version control enhances transparency and ensures reproducibility.
By leveraging scripting and sharing code, researchers can promote collaboration, verification, and efficient building upon each other's work in scientific research.
Getting Started¶
Jump into the codebases and our apps.
FAIR¶
The openDendro project is aligned with the FAIR data principles, emphasizing the Findability, Accessibility, Interoperability, and Reusability of research data. Our repository is designed to facilitate reproducible research, ensuring that data and software resources are organized, documented, and accessible. We prioritize interoperability, enabling seamless integration with existing tools and frameworks. By following open licensing and attribution practices, we promote the reusability of our resources, fostering replication, validation, and extension of research findings. Join us in our commitment to FAIR principles and advancing dendrochronology with open and transparent practices.



